
Radical Collaborations and Cornell Connections Power Systems Biology
by Nozomi Nishimura
(excerpts from Cornell Chronicle article "Facility upgrades invigorate immunology, cell research," by Elodie Smith, May 2, 2019.)
New data, tools and algorithms are driving the rapid rise of systems biology as a field, but Cornell's especially collaborative culture is the secret ingredient that accelerates discovery.
“Cornell has an excellent social atmosphere that invites collaboration, which is essential for work on complex systems,” said Ben Grodner, a Ph.D. student in the De Vlaminck Lab. Lauren Walter, a graduate student in the Cosgrove Lab, agrees. “There are a lot of resources available to systems biology students at Cornell, including faculty and students that are knowledgeable and passionate about their work. It is motivating to be around people willing to advise you when needed and to discuss research with you.”
Led by young faculty, Cornell BME is at the forefront of innovation in systems biology. Recently, BME assistant professors Ben Cosgrove and Iwjin De Vlaminck teamed up with Olivier Elemento, director of the Englander Institute for Precision Medicine at Weill Cornell Medicine, to an establish an ‘atlas’ of cells for muscle repair (see more in sidebar). The Cosgrove Lab studies the effects of aging on stem cell function and declining muscle regeneration using single cell technologies and systems bioengineering methods. Undergraduates also get to learn about these cutting-edge methods in Cosgrove’s popular class Cellular Systems (BME 3110), for which he was awarded the 2019 Dorothy G. Swanson Excellence in Teaching Award.
Other work from De Vlaminck shows how new measurement methods can directly impact patients. His lab has developed methods for extracting information from cell-free DNA that enables new diagnoses from easily attained samples like blood and urine. Using bioinformatic methods, De Vlaminck assembles DNA fragments to identify microbes. This method could replace traditional microbiology tests which take days to make cultures from clinical samples and are limited in the range of species that they detect. This technology has also been adapted to detect organ failure in a clinical application.
Assistant professor Ilana Brito also uses systems biology approaches to understand how the microbiome affects human health. Known for her work on horizontal gene transfer, Brito uses computational and experimental approaches to study how the microbiome evolves and interacts with its host. She also runs the Microbiome Hackathon, an immersive weekend experience that teaches about data science, the microbiome, and computational biology.
“At Cornell we have groups developing new imaging techniques, computational resources, and high-throughput molecular assays,” said David McKellar, a Ph.D. student jointly advised by Cosgrove and De Vlaminck. "What excites me most is the opportunity to combine each of these strategies to discover new biology.”
Mert Sabuncu, assistant professor of ECE and BME, facilitates these new scientific combinations. He is an innovator in machine learning with a focus on combining patient data such as MRI images and genetics data. While much of his work is on clinical data, he has recently expanded to using machine learning and image processing for analysis of experimental rodent studies with collaborators.
BME faculty are excited about several university-wide initiatives that expand systems biology capabilities. The university has invested heavily in new infrastructure to support this research direction. Hailed as “transformative” and “a historic achievement” by faculty members, a strategic investment of close to $2 million directed by Provost Michael Kotlikoff has improved Cornell’s capabilities in flow cytometry, which is pivotal in cell research.
(De Vlaminck Lab).
The investment from the provost’s office, along with significant contributions from the colleges of Veterinary Medicine, Engineering, Human Ecology, Agriculture and Life Sciences, and Arts and Sciences, allowed the purchase of four new flow cytometers. These new machines in addition to existing machines are housed in a new Flow Cytometry Core Facility, with sites in the biotechnology building and at the veterinary school. BME’s Cosgrove was central to the establishment of the new facility. “The new Flow Cytometry Facility is truly game-changing for the immunology community,” said Gary Koretzky ’78, Cornell’s vice provost for academic integration and inaugural director of the Center for Immunology.
Other new resources include an Epigenomics Core Facility that was established in 2020. The provost-led Radical Collaborations program includes genome and infection biology, providing resources to fund new collaborations across the university. These ensure that the systems biology projects born in Cornell’s friendly environment will have the latest tools and robust support.
Muscle stem cells compiled in ‘atlas’
By David Nutt for Cornell Chronicle
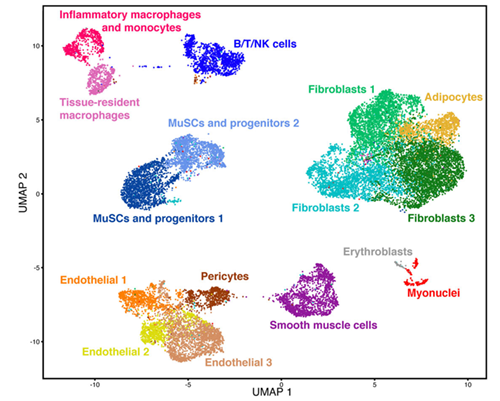
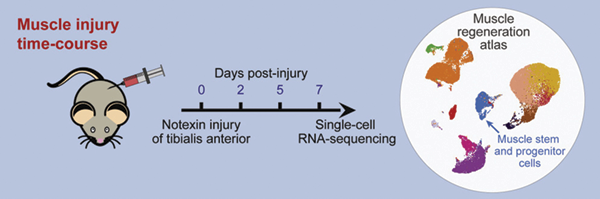
A team of Cornell researchers led by BME's Ben Cosgrove used a new cellular profiling technology to probe and catalog the activity of almost every kind of cell involved in muscle repair. They compiled their findings into a “cell atlas” of muscle regeneration that is one of the largest datasets of its kind. This resource provides a comprehensive picture of the many intricate cellular interactions in tissue self-repair and may potentially lead to better rehabilitation strategies and support for patients recovering from muscle injuries.
The team’s paper, “Single-Cell Analysis of the Muscle Stem Cell Hierarchy Identifies Heterotypic Communication Signals Involved in Skeletal Muscle Regeneration,” published March 10 in Cell Reports.
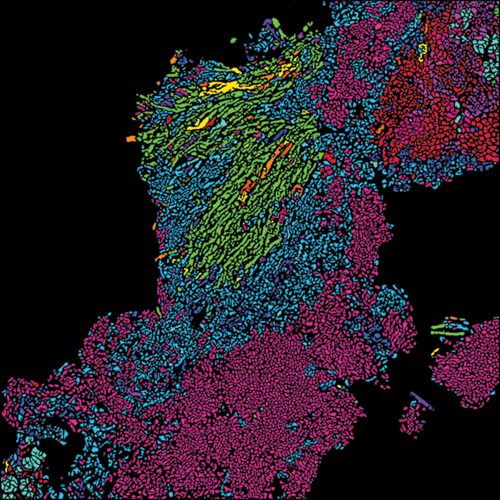
Cosgrove worked with Cornell’s Biotechnology Resource Center to use single-cell RNA sequencing to analyze the gene expression signatures in thousands of individual cells, all taken from the actively regenerating muscles of mice – including the rare muscle stem cells that drive the repair process. Cosgrove also collaborated with Iwijn De Vlaminck, the Meinig School's Robert N. Noyce Assistant Professor in Life Science and Technology. Together, they applied new algorithms to filter the extensive collection of molecular information.
Unlike a road atlas that uses spatial coordinates, this atlas is more of a “laundry list of the key cell players,” Cosgrove said, itemizing their similarities and differences, with 15 unique cell types in muscle tissue. This collection emphasizes how much heterogeneity exists within each cell type, a range of diversity that led the researchers to pursue the sources of specific molecular variations that might be otherwise overlooked.
“Now that we know more about what is happening in the healthy, normal adult repair process, we can ask ‘How do the cellular players get misallocated and misactivated in the disease settings?’” Cosgrove said.
Cosgrove’s group is currently applying this approach to look at muscle repair deficiency in aging and muscular dystrophy.
doi: https://doi.org/10.1016/j.celrep.2020.02.067

